Joseph K. Aicher

About
I am an MD-PhD student at University of Pennsylvania. I am currently completing my PhD in Genomics and Computational Biology co-advised by Drs. Yoseph Barash and Elizabeth Bhoj.
Scientifically, I am interested in machine learning, statistical modeling, genomics, and biomedical informatics, especially as they relate to human health. My current research focuses on developing and applying new computational tools and frameworks to improve diagnostic rates for people with suspected Mendelian disorders. I am developing new approaches to detect defects in RNA splicing from patient RNA-seq or to predict such changes directly from patient exome or genome sequencing.
During my PhD, I interned at Amazon and was awarded a Ruth L. Kirschstein National Research Service Award (F30) by the NIH for my research and training. Prior to starting my MD-PhD, I studied at University of Alabama for four years, receiving an M.A. in Mathematics and a B.S. in Mathematics and Physics. My master's thesis investigated the application of probabilistic graphical models to metabolomics data. I also spent two summers at University of North Carolina Chapel Hill creating computational physics simulations of the yeast mitotic spindle.
Email / CV / Google Scholar / LinkedIn
Publications
Lubin E,
Bryant L,
Aicher J,
Li D,
Bhoj E.
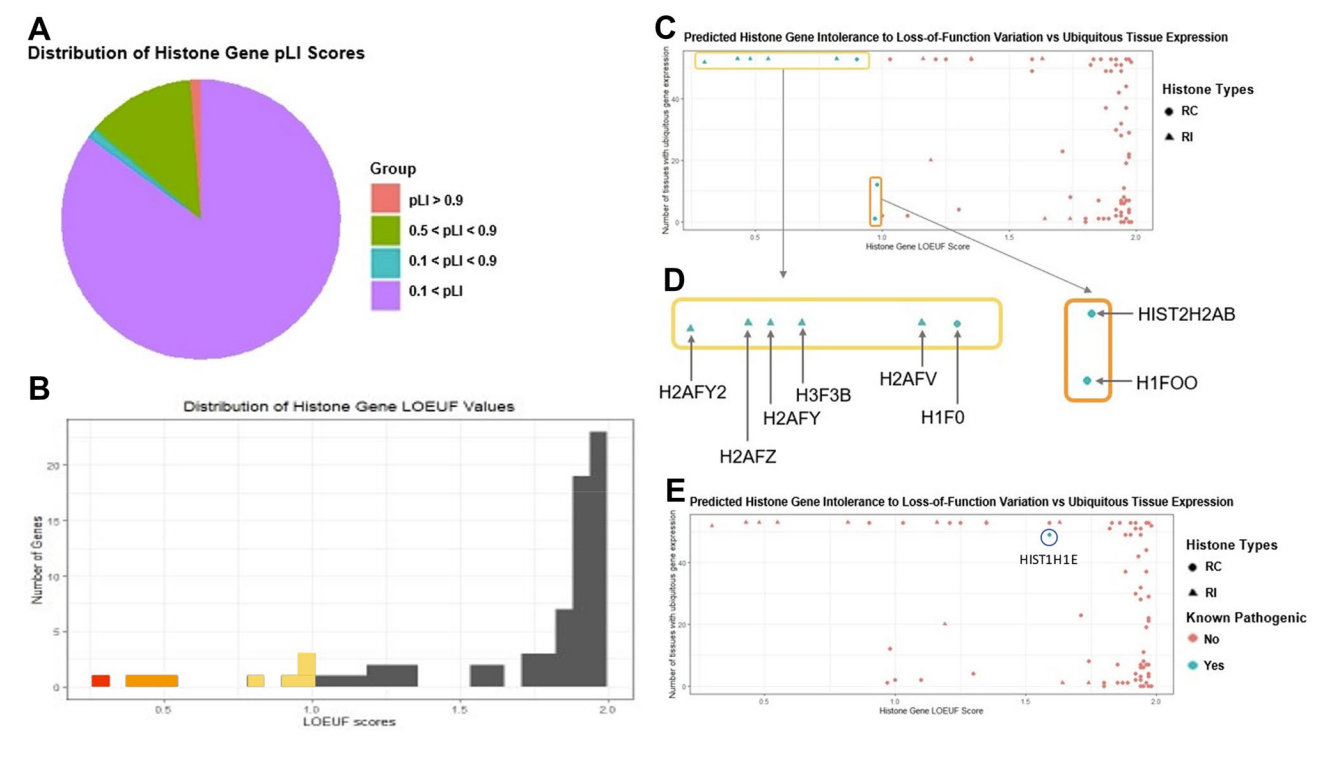
Analysis of histone variant constraint and tissue expression
suggests five potential novel human disease genes: H2AFY2, H2AFZ,
H2AFY, H2AFV, H1F0.
Human Genetics, 2022.
Jha A,
Aicher JK,
Gazzara MR,
Singh D,
Barash Y.
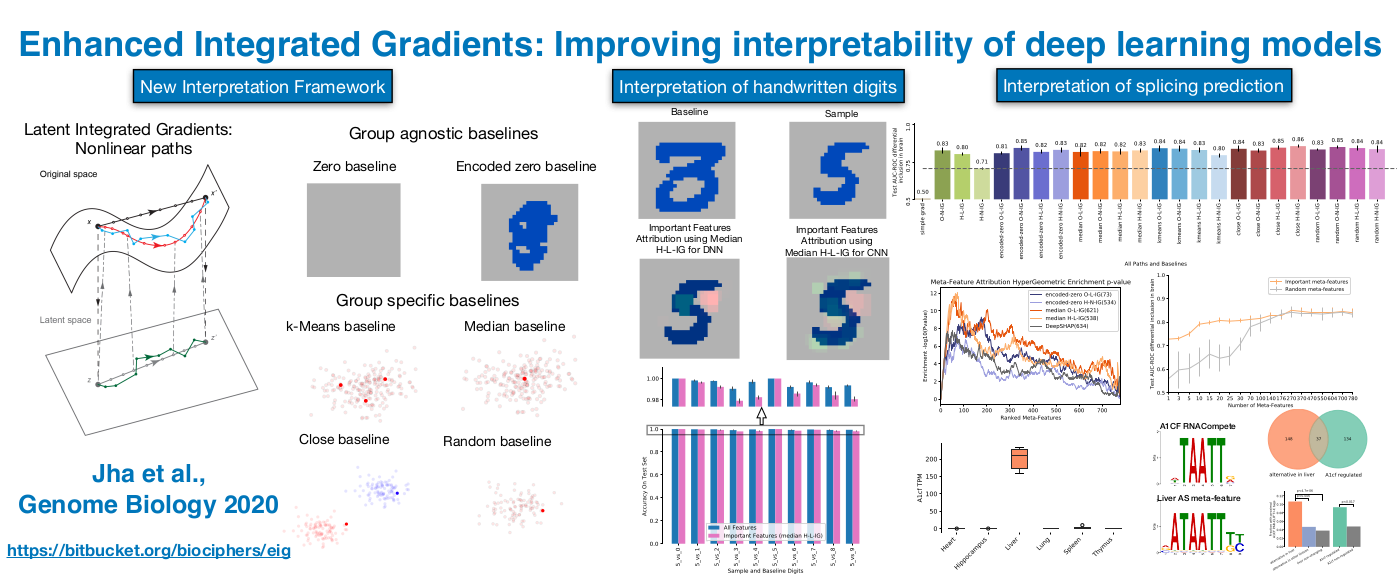
Enhanced Integrated Gradients: improving interpretability of
deep learning models using splicing codes as a case study.
Genome Biology, 2020.
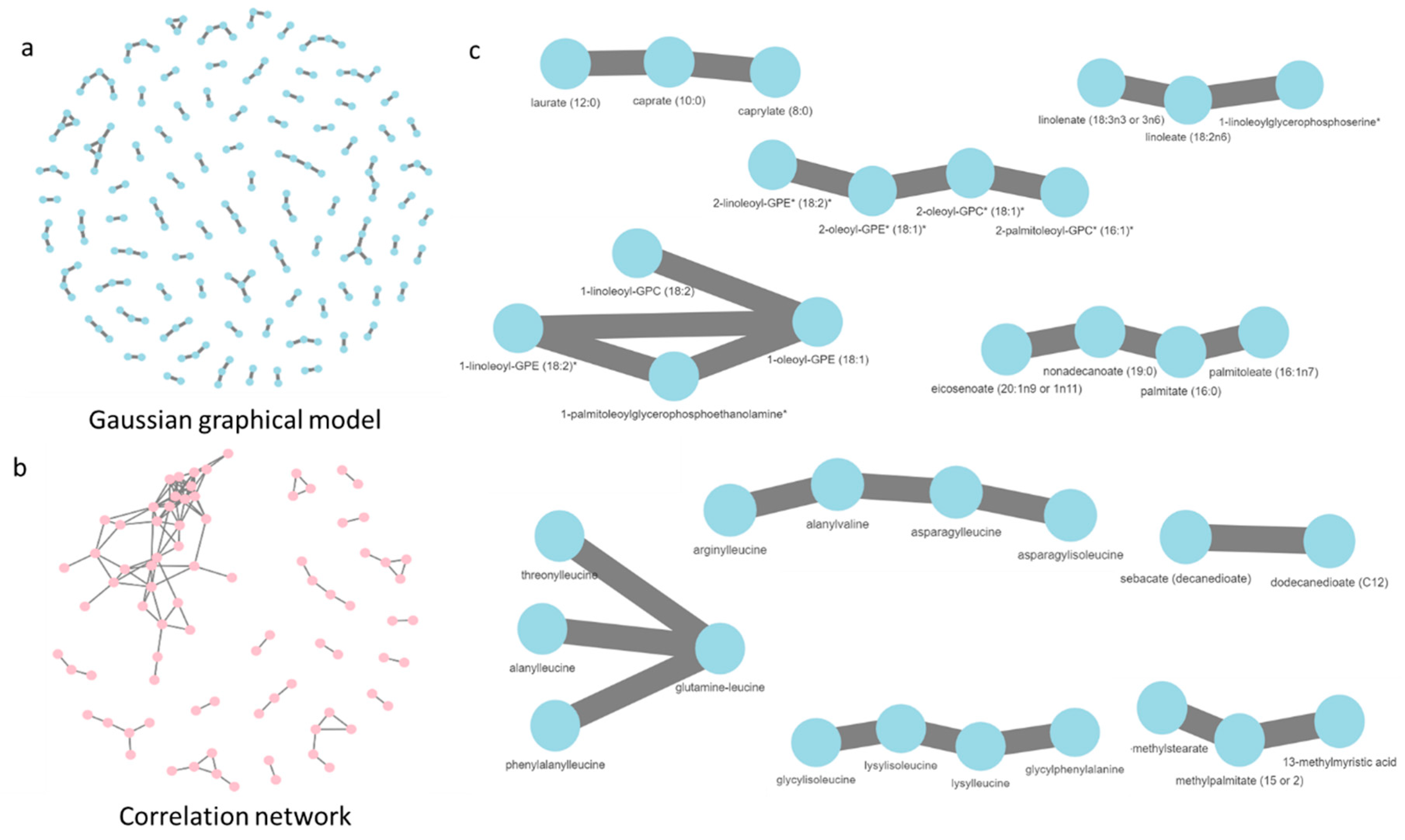
Oza VH,
Aicher JK,
Reed LK.
Random forest analysis of untargeted metabolomics data suggests
increased use of omega fatty acid oxidation pathway in
Drosophila melanogaster larvae fed a medium chain fatty acid
rich high-fat diet.
Metabolites, 2019.
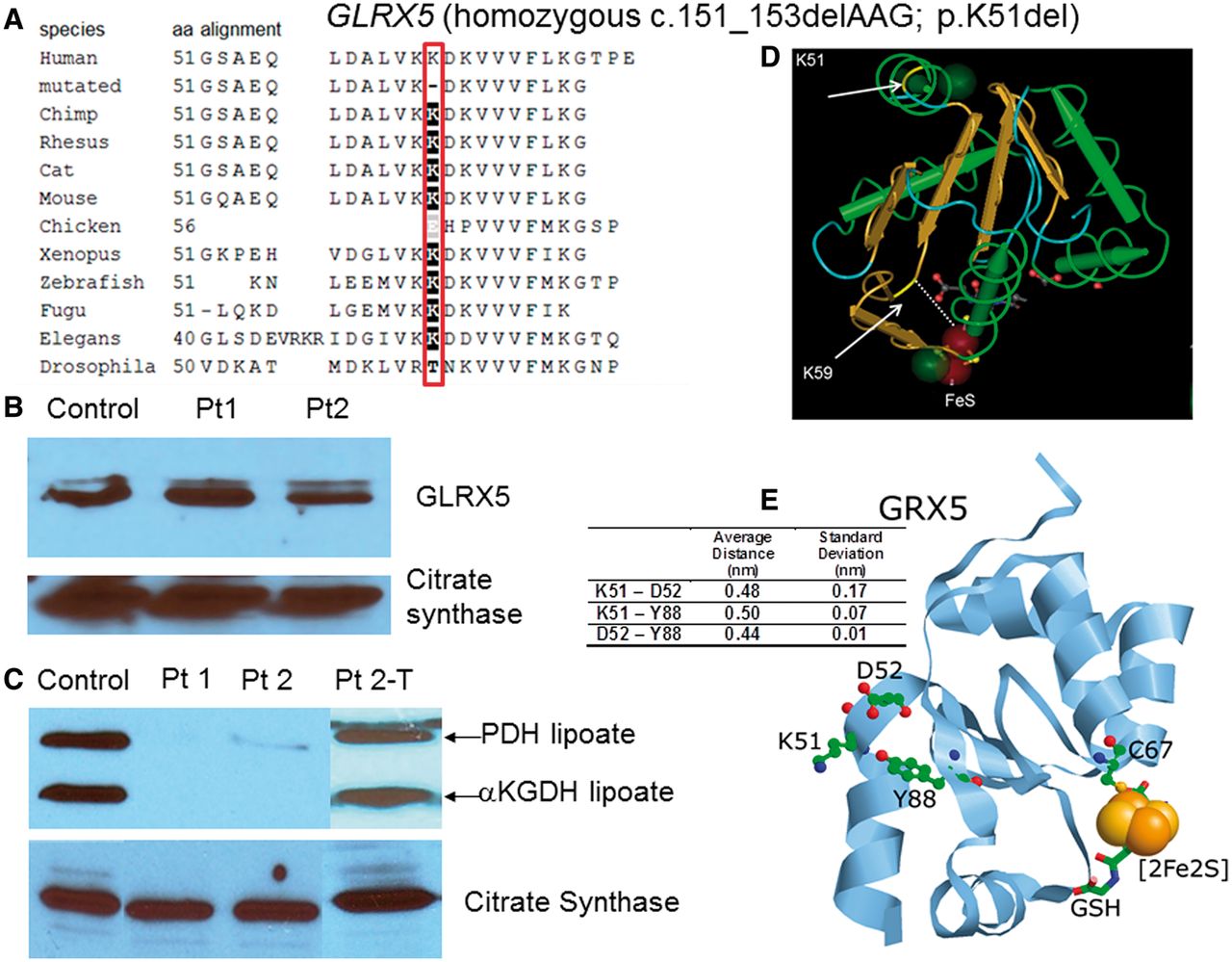
Baker PR,
Friederich MW,
Swanson MA,
Shaikh T,
Bhattacharya K,
Scharer GH,
Aicher J,
Creadon-Swindell G,
Geiger E,
MacLean KN,
Lee W-T,
Deshpande C,
Freckmann M-L,
Shih L-Y,
Wasserstein M,
Rasmussen MB,
Lund AM,
Procopis P,
Cameron JM,
Robinson BH,
Brown GK,
Brown RM,
Compton AG,
Dieckmann CL,
Collard R,
Coughlin CR,
Spector E,
Wempe MF,
Van Hove JLK.
Variant non ketotic hyperglycinemia is caused by mutations in LIAS, BOLA3 and the novel gene GLRX5.
Brain, 2014.